Superbug virulence regulatory mechanism revealed

As antibiotic resistance is growing and posing a threat on public health, developing new antibiotics has become more urgent than ever. Researchers at City University of Hong Kong (CityU) have recently revealed the virulence regulatory mechanism in Pseudomonas aeruginosa, a superbug which is common in patients with a weak immune system and is resistant to many antibiotics. The findings pave ways for identifying good antibiotic targets for new drug development.
Superbug Pseudomonas aeruginosa is a common pathogen of nosocomial infections, causing high morbidity and mortality in immunocompromised patients. It is also naturally tolerant to many clinically important antibiotics such as ampicillin, amoxicillin, and vancomycin. In 2017, the World Health Organization (WHO) classified this notorious bacterium as one of the three “critical priority pathogens” that new drugs are urgently needed.
A joint research led by Dr Deng Xin (Assistant Professor), a microbiologist, and Dr Wang Xin (Associate Professor), a computational biologist from Department of Biomedical Sciences (BMS) at CityU, have recently revealed the genomic regulatory network in Pseudomonas aeruginosa and identified the master regulators on key pathogenic pathways. The development of inhibitors against these newly identified master regulators can potentially lead to the discovery of novel drugs that target Pseudomonas aeruginosa.
The findings were published in the latest issue of Nature Communications, titled “An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa”.

Finding good antibiotic targets: master regulators
Bacterial pathogenicity is mediated via regulatory networks that include virulence-related transcriptional factors. Transcriptional factors (TFs) are proteins which can turn the specific genes (their functional target genes) “on” and “off”, generally a key determinant in whether the gene functions at a given time. And master regulators are the transcriptional factors that appear to control most of the regulatory activities of other transcriptional factors and the associated genes. Therefore, the master regulators are often good antibiotic targets that can be used for future drug development.
In Pseudomonas aeruginosa, numerous TFs regulate virulence by tuning quorum sensing (QS), the type III secretion (T3SS) and type VI secretion system (T6SS). In the past seven years, in collaboration with Professor Liang Haihua from Northwest University (China), Dr Deng has been working to reveal the pathogenesis of Pseudomonas aeruginosa, and to discover and clarify the regulation mechanism of multiple virulence-related TFs (Shao et al, J Bacteriol, 2018; Zhao et al, PLOS Biol, 2016; Kong et al, Nucleic Acids Res, 2015; Liang et al, Nucleic Acids Res, 2014; Liang et al, J Bacteriol, 2012).
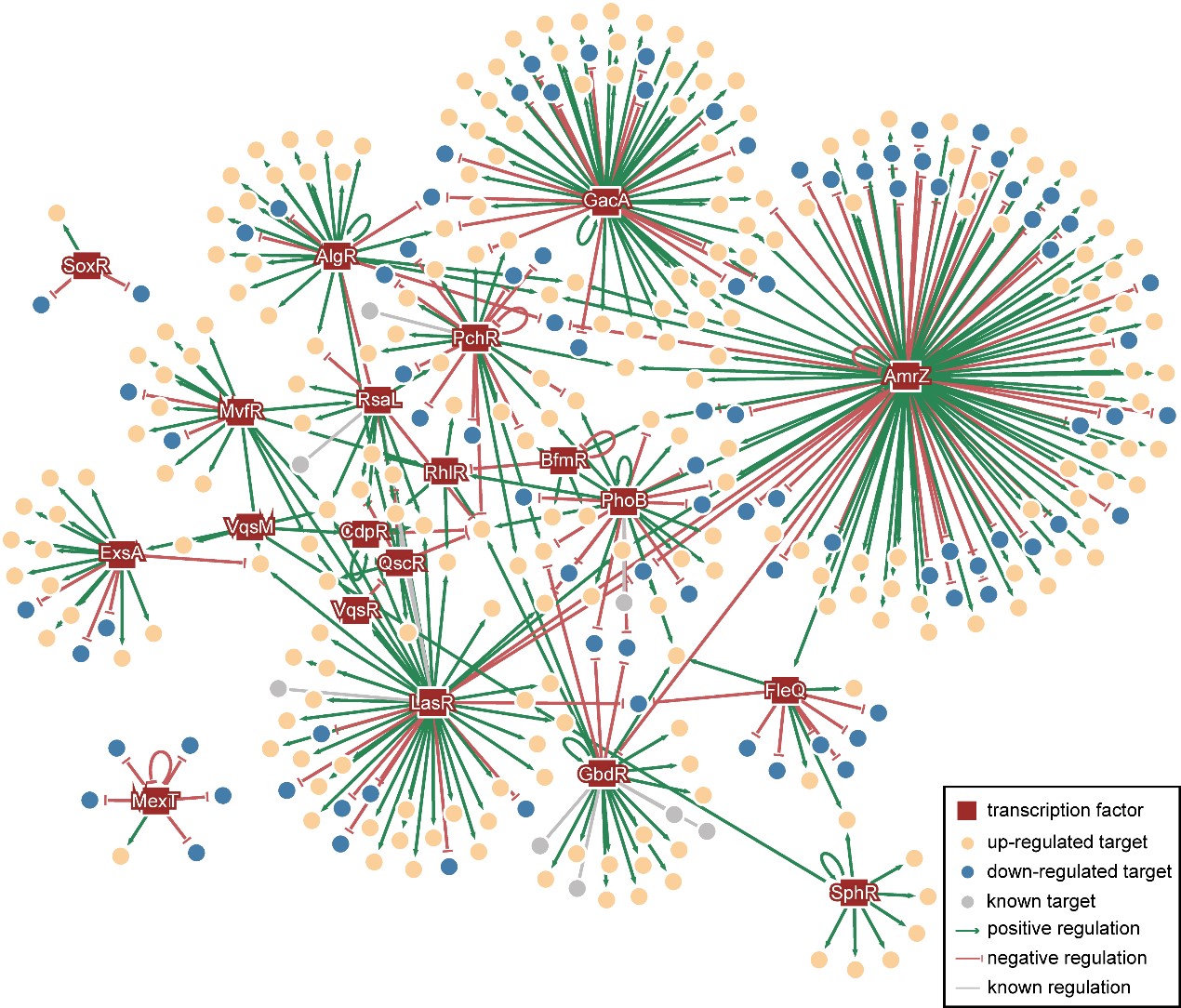
To further conduct a global analysis of the pathogenicity and discover new potential drug targets of Pseudomonas aeruginosa, Dr Deng's team and Dr Wang's team collaborated on the analysis and discovery of the crosstalk – signal pathway affecting another – in the known 20 virulence-related TFs. Subsequently, they mapped the Pseudomonas aeruginosa Genomic regulatory network (PAGnet) to encode the regulatory relationships of these 20 TFs with their functional target genes (Figure 1).
This PAGnet revealed the intricate mechanism of virulence regulation mediated by these TFs and the related crosstalk, and hence led to the identification of nine master regulators, in QS and T3SS.
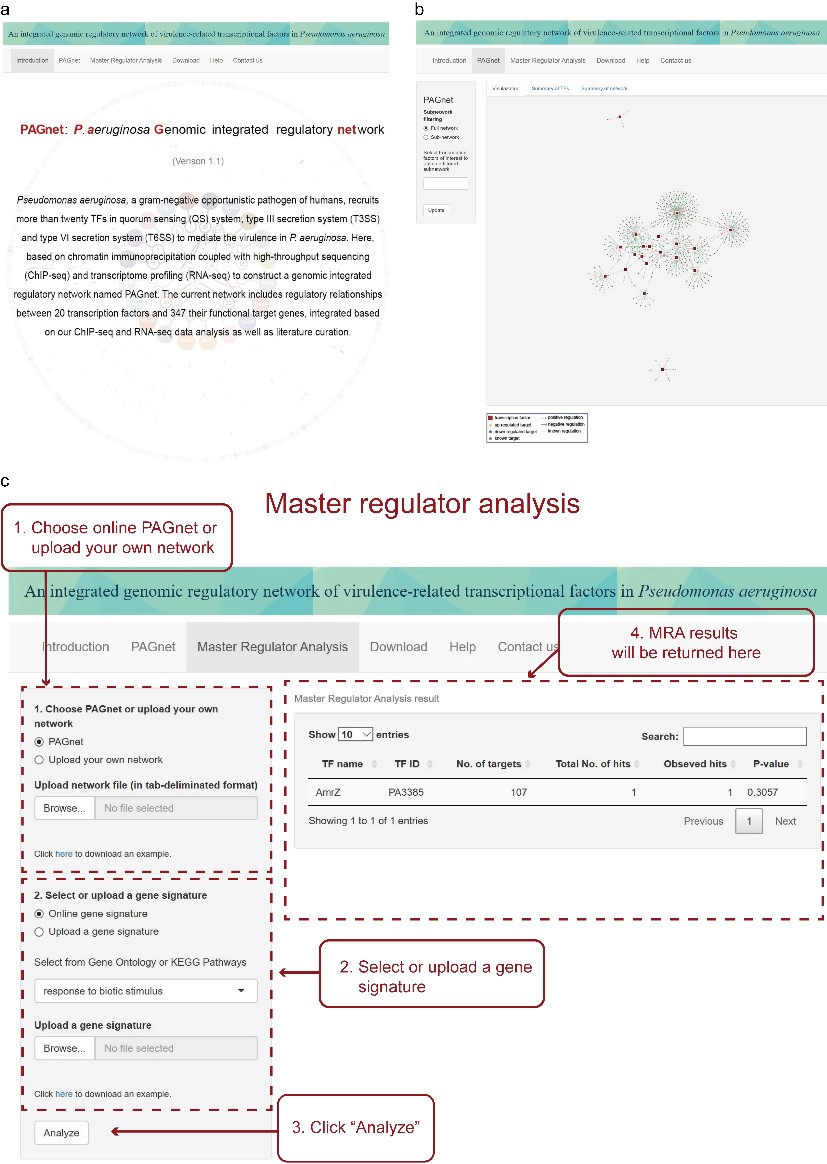
An online platform for potential wider pathological use
As a contribution to the research community, they have also developed an online platform (http://pagnetwork.org/) and R package (https://github.com/CityUHK-CompBio/PAGnet) based on PAGnet to ensure an up-to-date regulatory network and facilitate user-customized analyses (Figure 2). This platform and R package provide network visualization, subnetwork filtering and downloading services to the user, to facilitate the visualization and exploration of the virulence regulatory network, as well as master regulator analysis for the identification of key TFs that mediate a biological process or pathway in Pseudomonas aeruginosa.
“The master regulators we identified are potential antibiotic targets, which has important clinical significance for the development of new antibiotics for Pseudomonas aeruginosa in the future. More importantly, the network we build is not just for Pseudomonas aeruginosa, the methodology and conclusions of this work may be applicable to other bacterial pathogens in the future,” commented Dr Deng.
Dr Deng and Dr Wang are the correspondence authors of the paper. The first co-authors are PhD student Huang Hao, Xie Yingpeng and research assistant Dr Shao Xiaolong at CityU’s BMS Department. Other authors include PhD student Wang Tingting and research assistant Zhang Yingchao from the Department.
